Step 1. Retrieve the annotation and the sequence of the gene (EMBL database)
- Go to EMBL database at EBI
- mRNA sequence: Type U43653
in Nucleotide sequences
- On top, click over the EMBL:HS436531 entry
- Have a look at the description: IDs, references, attributes, sequences
- Search the Feature of Coding Sequence (FT CDS).
Click over and check the ORF correctness: the beginning and the end of the
sequence correspond respectively to the Start and Stop codons?
Step 2. Learn more about the Leptin gene
Using a genome browser
- Go back to the initial screen that contained the result of your
first query.
- On the left, you will find the Display
Options box.
- Select the FastaSeqs view and press
the button Apply Display Options
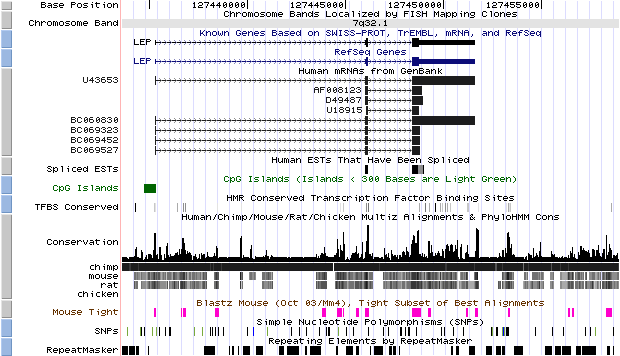
- Open the UCSC genome browser
- Select the alignment program Blat
(human genome)
- Paste the Fasta sequence of the Leptin gene and submit the query
- Browse the first hit in the list of matches
- Have a look at the different displaying options. We recommend to
zoom out 10x the initial picture to explore the genomic landscape around
the gene. For instance, try to:
- obtain the RefSeq gene sequence
- check the presence of a CpG island in the promoter
- examine the mRNAs supporting the gene annotation
- evaluate the conservation between orthologues
- Task1: What do you have to do if you want to see the computationally
predicted transcription factor binding sites?
- Task2: Try to locate the sequence in other genomes using BLAT (e.g. mouse)
Using the LocusLink database
- Go to LocusLink database at NCBI
- Type U43653
in Query
- Click on the entry LEP (leptin)
- Identify main fields in the entry: functional description, NM and NP
annotations
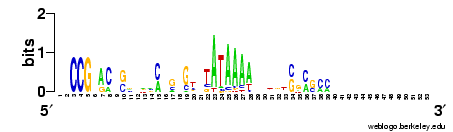
Step 3. PROMOTER information: sequence and experimental annotation
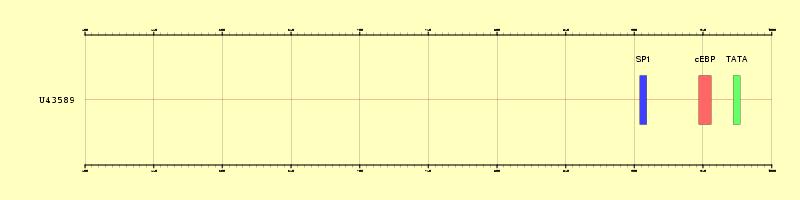 |
|
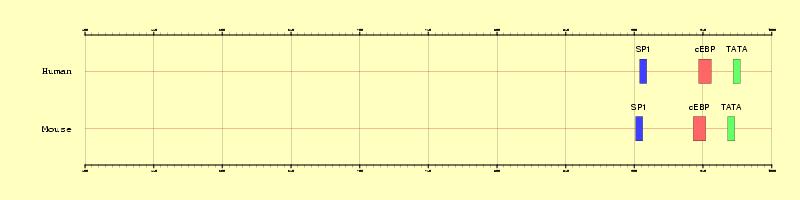
Figure 1. Graphical representation of the three regulatory
elements annotated in the promoter U43589 (500 bps upstream the TSS)
|
|
- J.F. Abril and R. Guigó.
gff2ps: visualizing genomic annotations.
Bioinformatics 16:743-744 (2000).
-
Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E.
TRANSFAC: transcriptional regulation, from patterns to profiles.
Nucleic Acids Research 31:374-378 (2003).
-
van Helden J. Regulatory sequence analysis tools.Nucleic
Acids Res. 31:3593-3596 (2003).
-
JD Thompson, DG Higgins, and TJ Gibson. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acid Res. 22:4673-4680 (1994).
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215:403-410 (1990).
-
Timothy L. Bailey and Charles Elkan.
Fitting a mixture model by expectation maximization to discover motifs in biopolymers.
Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California (1994).
-
Roth, FR, Hughes, JD, Estep, PE & GM Church. Finding DNA Regulatory Motifs
within Unaligned Non-Coding Sequences Clustered by Whole-Genome mRNA
Quantitation. Nature Biotechnology 16:939-945 (1998).
- X. Messeguer, R. Escudero, D. Farré, O. Núñez, J. Martínez and M.Mar Albà.
PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics Vol. 18: 333-334 (2002).
-
Mason MM, He Y, Chen H, Quon MJ, Reitman M. Regulation of leptin promoter function by Sp1, C/EBP, and a novel factor. Endocrinology. 139:1013-1022 (1998).
|