| |
To build a model or representation of the family of binding sites that are known to associate
a given TF, it is very common to put together the examples, then align them and finally create
a weight matrix or a sequence logo to detect the conserved positions.
With ABS, the users can retrieve the set of motifs annotated as binding sites for a certain TF.
Additionally, the sites can be grouped by species to detect conservations that are specific of
some species.

The output consists of three parts:
I. The binding sites of ABS associated to this TF (of this species)
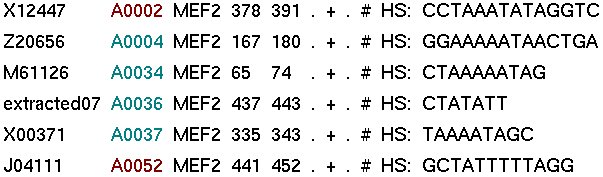
|
-> There is a link to access directly the gene entry in which each binding site is annotated
|
II. The alignment of the sites

|
III. The sequence logo

|
| | |
CopyRight © 2005
ABS is under GNU General Public License.
|
|