| |
To evaluate the accuracy of a motif search program is necessary to compare the predictions
that this program produces in a set of annotated sequences with the real coordinates
of the motifs on such sequences.
For instance, the real motifs here are located in a real promoter sequence:
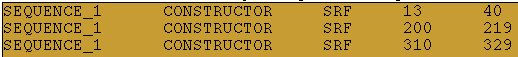
And these could be the predictions of a pattern discovery program:
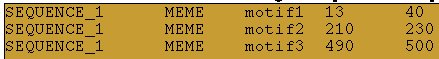
You can see that:
- The predicted motif1 is perfectly supported by a real annotation
- The predicted motif2 is partially supported by a real annotation
- The third real site is not detected in the predictions
- The predicted motif3 is not supported by any real data
Graphically, the comparison is easier:
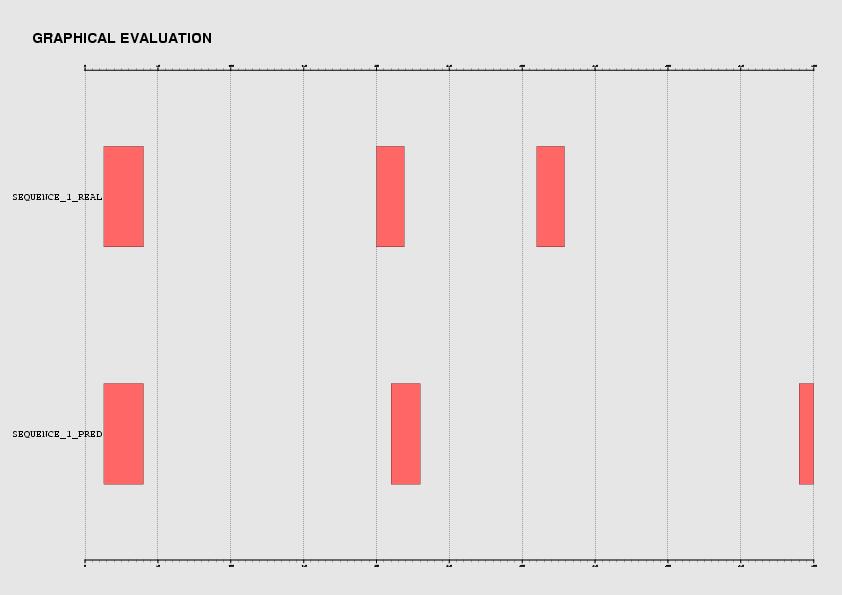
In the output of the Evaluator program, a set of typical accuracy measures are displayed.
Essentially, these values are useful to measure the sensitivity to capture the real
sites and the specificity to avoid artifacts in the predictions.
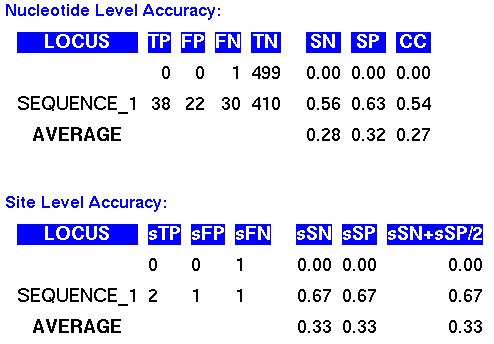
A complete summary of the accuracy values is included within the output.
For further information, check the following references:
- Burset, M. and Guigo, R. Evaluation of gene structure prediction programs. Genomics 34: 353-367 (1996).
- Tompa et al. Assessing computational tools for the discovery of transcription factor binding sites. Nature Biotechnology 23: 137-144 (2005).
CopyRight © 2005
ABS is under GNU General Public License.
|
|